Research Interests
[1] Structural Understanding of Ion Channel Action, Assembly, and Regulation
Propagation of electrical signals in the cardiac and nervous systems requires the concerted action of ion channel proteins and the molecules that modulate their activity. Understanding ion channels ultimately requires a high-resolution structural description of the channel proteins, their assembly process, regulatory factors, and conformational changes that accompany channel action. Our lab approaches this problem from the perspective of structural biology. Because channels are membrane proteins, a difficult class to investigate with any single structural technique, our efforts are directed at a multidisciplinary approach that involves a breadth of methods such as cryo-electronmicroscopy, X-ray crystallography, chemical approaches, and genetics for studying ion channel structure and function. Our current research aims to understand the central types of channels that are responsible for excitation (voltage-gated calcium channels and voltage-gated sodium channels) and inhibition (potassium channels) of electrical activity.
Key Publications
EMC chaperone-CaV structure reveals an ion channel assembly intermediate. Chen Z, Mondal A, Aberemane-Ali, F, Jang S, Niranjan S, Montaño JL, Zaro BW, Minor, D.L., Jr. Nature 619 410-419 (2023)
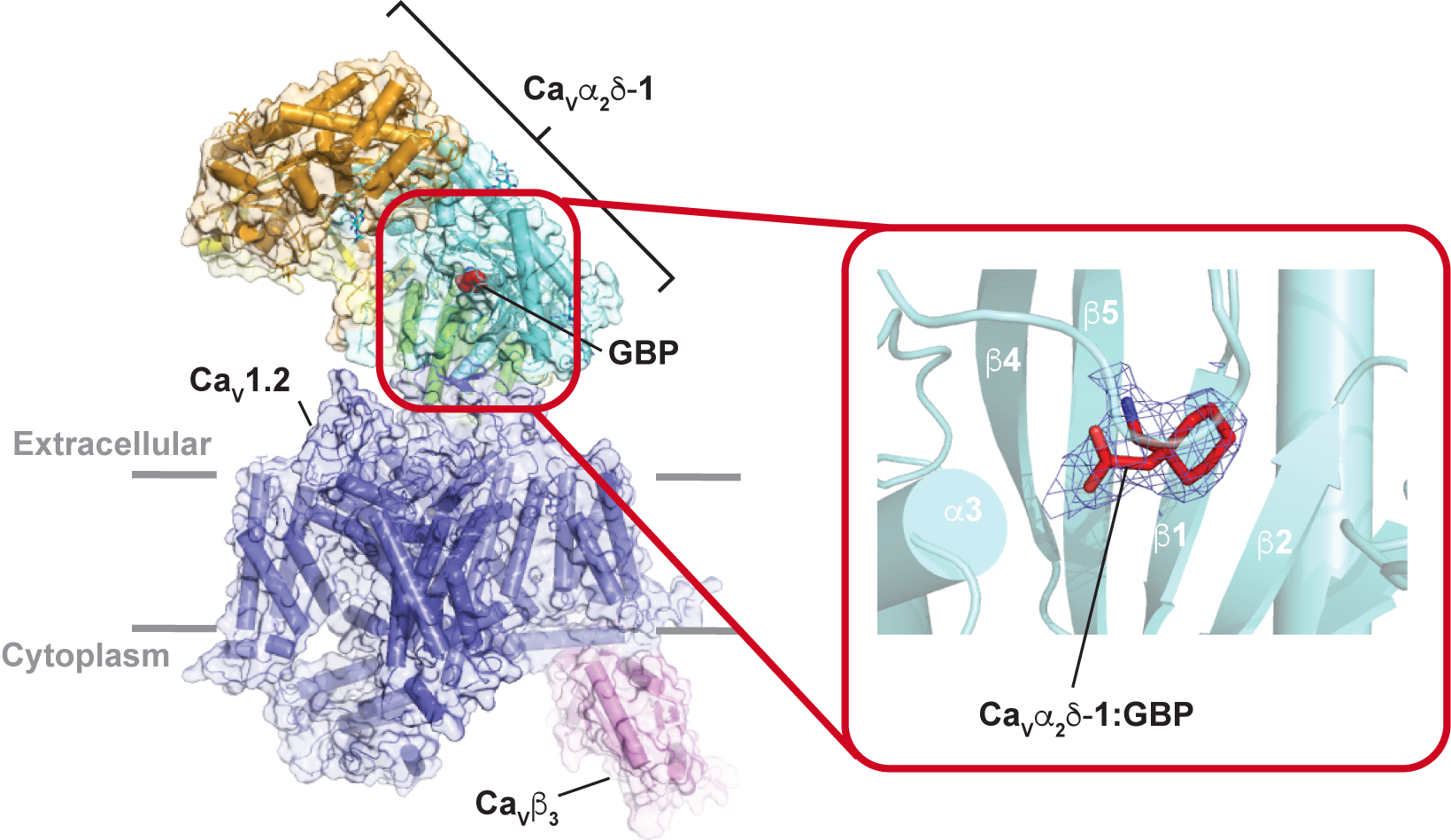
Structural basis for CaVɑ 2δ:gabapentin binding. Chen Z, Mondal A, and Minor, D.L., Jr. Nature Struct. & Mol. Biol. 30 735-739 (2023)
Additional Press
» UCSF Newscenter Article
» Nature Structure and Molecular Biology News & Views
Quaternary structure independent folding of voltage-gated ion channel pore domain subunits. Arrigoni, C., Lolicato, M., Shaya, D., Rohaim, A., Findeisen, F., Fong L.-K., Colleran, C.M., Dominik, P., Kim, S.S., Schuermann, J., DeGrado, W.F., Grabe, M., Kossiakoff, A.A., and Minor, D.L. Jr., Nature Structural and Molecular Biology 29 537-548 (2022)
Structural insights into the mechanisms and pharmacology of K2P potassium channels. Natale, A.M., Deal, P.E., and Minor, D.L. Jr., Journal of Molecular Biology 433:166995 (2021) PMID: 33887333 PMCID: PMC8436263
Production of K2P2.1 (TREK-1) for structural studies. Lee, H., Lolicato, M., Arrigoni, C., Minor, D.L. Jr., Methods in Enzymology 653:151-188. PMID: 34099170. (2021)
K2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions. Lolicato, M., Natale, A.M., Abderemane-Ali, F., Crottes, D., Capponi, S., Duman, R. Wagner, A., Rosenberg, J.M., Grabe, M., Minor, D.L. Jr., Science Advances 6 eabc9174 (2020)
A selectivity filter gate controls voltage gated calcium channel (CaV) calcium-dependent inactivation. Abderemane-Ali, F., Findeisen, F., Rossen, N.D., and Minor, D.L. Jr., Neuron 101 1134-1149 (2019) PMID: 30733149 PMCID.
K2P2.1(TREK-1):activator complexes reveal a cryptic selectivity filter binding site. Lolicato, M., Arrigoni, C., Mori, T., Sekioka, Y., Bryant, C., Clark, K.A., Minor, D.L., Jr., Nature 547 364-368 (2017) PMID: 28693035 PMCID: PMC5778891
Unfolding of a temperature-sensitive domain controls voltage-gated channel activation. Arrigoni, C., Rohaim, A., Shaya, D., Findeisen, F., Stein, R.A. Nurva, S.R., Mishra, S., Mchaourab, H.S., and Minor, D.L., Jr., Cell 164 922‑936 (2016) PMID: 26919429 PMCID:PMC4769381
Transmembrane helix straightening and buckling underlies activation of mechanosensitive and thermosensitive K2P channels. Lolicato, M., Riegelhaupt, P.M., Arrigoni, C., Clark, K.A., Minor, D.L., Jr., Neuron 84 1198‑1212 (2014) PMID: 25500157 PMCID: PMC4270892
Multiple modalities act through a common gate to control K2P channel function. Bagriantsev, S., Clark, K.A., Peyronnet, R., Honoré, E., and Minor, D.L., Jr., The EMBO Journal 30 3594-3606 (2011)
Structure of a complex between a voltage‑gated calcium channel β-subunit and an α-subunit domain. Van Petegem, F., Clark, K.A., Chatelain, F.C., and Minor, D.L., Jr., Nature 429 671-675 (2004) PMID:15141227
[2] Ion Channel Chemical Biology
A rich diversity of ion channel genes and families are now known from molecular cloning and genome sequencing efforts. This abundance of ion channel genes poses dual problems: most have unknown functions and few have defined pharmacologies that can be used to dissect their biophysical or physiological mechanisms of action. Indeed, there are entire channel families that remain effectively pharmacological orphans. Unraveling the functions of this multitude of ion channels demands the development of new chemical biology tools that can be used to control the action of a given ion channel in both in vivo and in vitro settings.
We are pursuing a range of approaches to create and develop new protein-based and small molecule-based ion channel modulators that will expand the repertoire of novel ion channel-directed chemical biology agents. Our efforts deploy genetic selection methods to generate high affinity, specific inhibitors and activators of ion channels through the use of molecular evolution and selection methods (Bagriantsev et al. 2013, 2014), structure-based design of protein-protein interaction inhibitors (Findeisen et al., 2017), and structure-based development of ion channel small molecule modulators (Lolicato, et. al. Nature, 2017, Pope et al. ACS Chemical Neuroscience, 2018, Lolicato et al. Science Advances 2020, Pope et al. Cell Chemical Biology, 2020). These new tools should aid in unlocking unlock a wide range of currently unaddressable questions ranging from basic channel biophysics to physiology.
Key Publications
Differential effects of modified batrachotoxins on voltage-gated sodium channel fast and slow inactivation. MacKenzie, T.M.G., Abderemane-Ali, F., Garrison, C.E., Minor, D.L., Jr., Du Bois, J. Cell Chemical Biology Dec 24;S2451-9456(21)00517-1. doi:10.1016/j.chembiol.2021.12.003 (2021)
K2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions. Lolicato, M., Natale, A.M., Abderemane-Ali, F., Crottes, D., Capponi, S., Duman, R. Wagner, A., Rosenberg, J.M., Grabe, M., Minor, D.L. Jr., Science Advances 6 eabc9174 (2020)
Polynuclear Ruthenium Amines Inhibit K2P Channels via a "Finger in the Dam" Mechanism. Pope, L., Lolicato, M., Minor, D.L. Jr., Cell Chemical Biology 27, 511-524 (2020)
Protein and chemical determinants of BL-1249 action and selectivity for K2P channels. Pope. L., Arrigoni, C., Lou, H., Bryant, C. Gallardo-Gadoy, A., Renslo, A.R., and Minor, D.L. Jr., ACS Chemical Neuroscience 9 3153-3165, (2018) PMID: 30089357 PMCID: PMC6302903
K2P2.1(TREK-1):activator complexes reveal a cryptic selectivity filter binding site. Lolicato, M., Arrigoni, C., Mori, T., Sekioka, Y., Bryant, C., Clark, K.A., Minor, D.L., Jr., Nature 547 364-368 (2017) PMID: 28693035 PMCID: PMC5778891
Stapled voltage-gated calcium channel (CaV) α-Interaction Domain (AID) peptides act as selective protein-protein interaction inhibitors of CaV Function. Findeisen, F., Campiglio, M., Jo, H., Abderemane-Ali, F., Rumpf, C.H., Pope, L., Rossen, N.D., Flucher, B.E., DeGrado, W.F., and Minor D.L., Jr., ACS Chemical Neuroscience 8 1313-1326 (2017) PMID: 28278376 PMCID: PMC5481814
Tethered protein display identifies a novel Kir3.2 (GIRK2) regulator from protein scaffold libraries. Bagriantsev, S.N., Chatelain, F.C., Clark, K.A., Alagem, N., Reuveny, E., Minor, D.L., Jr., ACS Chemical Neuroscience 5 812‑822 (2014) PMID: 25028803 PMCID: PMC4176385
A high-throughput functional screen identifies small molecule regulators or temperature- and mechano-sensitive K2P channels. Bagriantsev, S. N., Ang, K.H., Gallardo-Godoy, A, Clark, K.A., Arkin, M.R., Renslo, A.R, and Minor, D.L., Jr,. ACS Chemical Biology 8 1841-1851 (2013) PMID: 23738709PMCID: PMC3747594
[3] Molecular mechanisms of toxin resistance
Ion channels are the targets of many naturally occurring small molecule toxins such as saxitoxin (STX), the paralytic agent produced by oceanic ‘red tide’ blooms, and the poison dart frog toxin batrachotoxin (BTX). The mechanisms by which animals that carry or regularly encounter such toxins resist poisoning remain poorly understood. In parallel with our interest in ion channels, we seek to uncover the molecular mechanisms of toxin resistance conferred by toxin binding proteins that act as ‘molecular sponges’ that neutralize toxins such as STX by sequestration (Abderemane-Ali et al. J. Gen. Physiol. 2021). We have a special interest in the saxiphilin family of amphibian STX binding proteins (Yen et al. Science Advances, 2019, Chen et al. PNAS 2022). Understanding the basic rules for how proteins recognize STX, BTX, and other toxins should aid in the development of new means to detect and neutralize such toxins.
Key Publications
Definition of a saxitoxin (STX) binding code enables discovery and characterization of the Anuran saxiphilin family. Chen, Z., Zakrzewska, S., Hajare, H., Alvarez-Bullya, A., Abderemane-Ali, F., Bogan, M., Ramirez, D., O’Connell, L.A., Du Bois, J., and Minor, D.L., Jr., Proceedings of the National Academy of Sciences, USA 119:e2210114119 (2022)
Additional Press
» Advanced Photon Source, Argonne National Lab, Science Highlight, 2023
Evidence that toxin resistance in poison birds and frogs is not rooted in sodium channel mutations and relies on ‘toxin sponge’ proteins. Abderemane-Ali, F., Rossen, N.D., Kobiela, M.E., Craig, R.A.II, Garrison, C.E., Chen, Z., O'Connell, L.A., Du Bois, J., Dumbacher, J.P., and Minor, D.L. Jr., Journal of General Physiology 153:e202112872 (2021) PMID: 34351379 PMCID: PMC8348241
Additional Press
» UCSF Newscenter Article
» Nature Highlights
» National Geographic
» The Times (UK)
» Live Science Article
Sodium channel toxin-resistance mutations do not govern batrachotoxin (BTX) autoresistance in poison birds and frogs. Abderemane-Ali, F., Rossen, N.D., Kobiela, M.E., Craig, R. A.II, Garrison, C.E., O’Connell, L.A., Du Bois, J., Dumbacher, J.P., and Minor, D.L., Jr.
Preprint: https://www.biorxiv.org/content/10.1101/2020.10.29.361212v1
Structure of the saxiphilin:saxitoxin (STX)complex reveals a convergent molecular recognition strategy for paralytic toxins. Yen, T.-J., Lolicato, M., Thomas-Tran, R., Du Bois, J., and Minor, D.L. Jr., Science Advances 5 eaax2650 (2019) PMID: 31223657 PMCID: PMC6584486
Additional Press
» Berkeley Lab Newscenter Article
» KQED Article
» UCSF Article
» Hakai Magazine Article